Approaches towards insights on pathogen evolution
Key Points
- Microbial genomics leverages high-throughput sequencing techniques to study genomic sequences of microorganisms.
- Ability to provide comprehensive insights into pathogen identity, diversity, and evolution is critical in accelerating vaccine development for human and animal health.
- Bioinformatics workflows to implement approaches such as phylogenetic analysis and taxonomic classification are key to generating valuable insights.
Background: Importance of microbial genomics
Microbial genomics is a high-throughput OMICS-based technique that entails the study of the genomic sequences of microorganisms. Research in microbial genomics has provided us with many insights on microbiome functioning, and refinements to perturbation methods to ultimately improving both human and animal health.1
The ability to provide comprehensive insights on pathogen identity, diversity, and evolution is invaluable in modern infectious disease research, and has profound implications for the development of vaccines.
Challenges in analyzing microbial genomics data for vaccine development
Due to the several benefits of OMICS-based technologies, analysis and interpretation of omics data for understanding the spread and evolution of pathogens is increasingly important for vaccine development. As more and more variety of omics data is being generated, a number of tools have been developed to process the data from quality checking to downstream analysis. This brings both experimental as well as computational challenges in acquisition and analysis of data2. Thus, we urgently need bioinformatics workflows that leverage the existing tools and are tailored to business requirements.
Approaches to generate insights on pathogen evolution:
Here, we summarize a few approaches to analyze the pathogen evolution from microbial genomics data and highlight our experience and expertise in implementing them in workflows.
- Phylogenetic analysis:
- This technique is used for studying evolutionary relatedness among various groups of organisms. A traditional workflow would involve multiple sequence alignment tool such as MAFFT, MUSCLE, ClustalW followed by phylogenetic tree building tool such as PHYLIP, FastTree.
- Recently we leveraged NextStrain3, an open-source tool for pathogen evolution, to develop a custom solution for a large pharma customer. The solution helped our customer to explore and visualize publicly available data alongside company-internal data. It was deployed in company’s environment so that the business users can freely and securely utilize it. Figure 1 shows a visualization of publicly available SARS-CoV-2 data on Nextstrain’s dashboard.
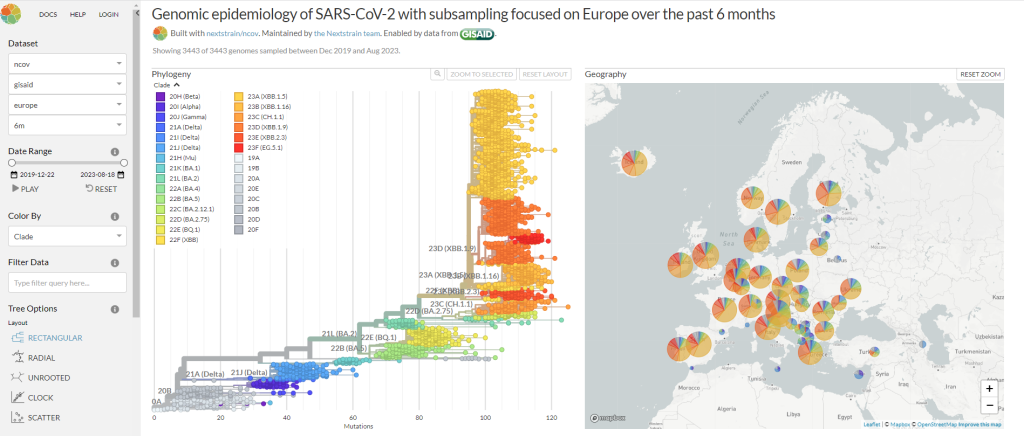
Figure 1: NextStrain’s dashboard based on publicly available SARS-COV data (Adapted from https://nextstrain.org/ncov/global)
- Taxonomic classification:
- One approach to analyze novel pathogens is to perform assignments of sequences to taxonomic groups based on sequence similarity. Tools such as Kraken2 work by leveraging exact k-mer matches between input sequence and a database containing reference sequences with taxonomic information.
- We implemented a previously published workflow4 to automate the analysis of multiple runs containing genomic sequences of animal samples from raw reads to interactive visualization for classification of novel pathogens. The workflow filters high quality reads and efficiently reports confidence scores for the classification results. With this workflow, our customer could generate insights on novel pathogens at a much higher pace than before, and could accelerate the vaccine development by focusing on pathogens-of-interest in the downstream validation processes. An exemplary report is provided in figure 2, which depicts the classification of sequences found in a human gut sample.
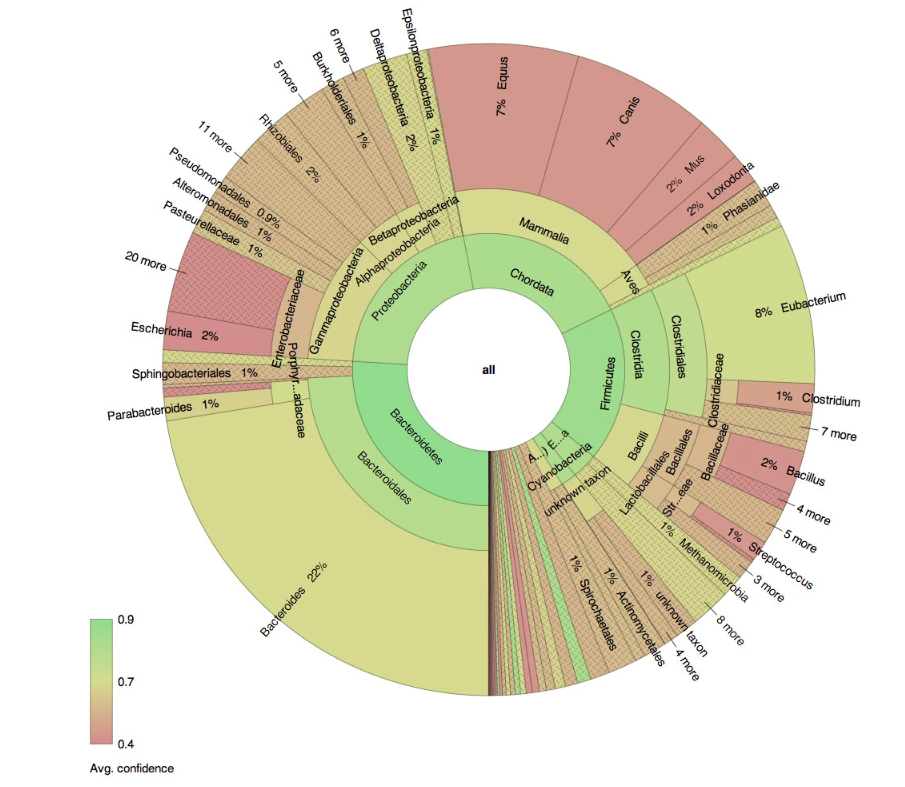
Figure 2: Exemplary interactive visualization performed on microbial genomics data (Source: Interactive visualization of taxonomic classification in Krona)
Our expertise and learnings to boost vaccine development initiatives
Figure 3: Our expertise and learnings to boost vaccine development initiatives
How can OSTHUS support you in your vaccine development initiatives?
- We have Bioinformatics domain knowledge to interpret existing datasets and provide support on its relevancy and better impact based on literature.
- We provide technology agnostic scientific advisory and data management solutions from vision and strategy, to market analysis, to implementation.
- We develop and/or automate customized workflows that not only fit into existing tool ecosystem but also scale to meet the dynamic data processing demands in terms of compute, storage, performance etc.
- We can spearhead the development of standardized workflows and best practices for reuse in order to avoid siloed solutions within the company.
Contact us to get the details on how we are helping our customers in accelerating their vaccine development programs.
References:
- Current challenges and best-practice protocols for microbiome analysis
Challenges in Bioinformatics Workflows for Processing Microbiome Omics Data at Scale- Nextstrain: real-time tracking of pathogen evolution
Disclaimer
The contents of this blog are solely the opinion of the author and do not represent the opinions of PharmaLex GmbH or its parent Cencora Inc. PharmaLex and Cencora strongly encourage readers to review the references provided with this blog and all available information related to the topics mentioned herein and to rely on their own experience and expertise in making decisions related thereto.





